The Polymerase Chain Reaction (Pcr) Can Be Used to Produce __________ of Copies in a Few Hours.
Chapter 10: Introduction to Biotechnology
10.1 Cloning and Genetic Engineering
Learning Objectives
Past the terminate of this section, yous will be able to:
- Explain the basic techniques used to dispense genetic cloth
- Explain molecular and reproductive cloning
Biotechnology is the utilise of artificial methods to modify the genetic material of living organisms or cells to produce novel compounds or to perform new functions. Biotechnology has been used for improving livestock and crops since the starting time of agriculture through selective convenance. Since the discovery of the structure of DNA in 1953, and particularly since the development of tools and methods to manipulate DNA in the 1970s, biotechnology has get synonymous with the manipulation of organisms' DNA at the molecular level. The main applications of this technology are in medicine (for the production of vaccines and antibiotics) and in agriculture (for the genetic modification of crops). Biotechnology also has many industrial applications, such as fermentation, the treatment of oil spills, and the production of biofuels, as well as many household applications such as the use of enzymes in laundry detergent.
Manipulating Genetic Material
To reach the applications described above, biotechnologists must be able to extract, manipulate, and clarify nucleic acids.
Review of Nucleic Acid Structure
To sympathise the basic techniques used to work with nucleic acids, remember that nucleic acids are macromolecules made of nucleotides (a saccharide, a phosphate, and a nitrogenous base). The phosphate groups on these molecules each have a cyberspace negative charge. An entire set of Deoxyribonucleic acid molecules in the nucleus of eukaryotic organisms is called the genome. Deoxyribonucleic acid has ii complementary strands linked by hydrogen bonds between the paired bases.
Unlike Dna in eukaryotic cells, RNA molecules get out the nucleus. Messenger RNA (mRNA) is analyzed most oft considering it represents the poly peptide-coding genes that are being expressed in the cell.
Isolation of Nucleic Acids
To study or manipulate nucleic acids, the DNA must starting time be extracted from cells. Various techniques are used to extract different types of Deoxyribonucleic acid (Figure 10.2). Nigh nucleic acid extraction techniques involve steps to intermission open the cell, and and so the employ of enzymatic reactions to destroy all undesired macromolecules. Cells are broken open using a detergent solution containing buffering compounds. To prevent degradation and contagion, macromolecules such as proteins and RNA are inactivated using enzymes. The DNA is then brought out of solution using alcohol. The resulting Dna, considering it is made upwards of long polymers, forms a gelled mass.
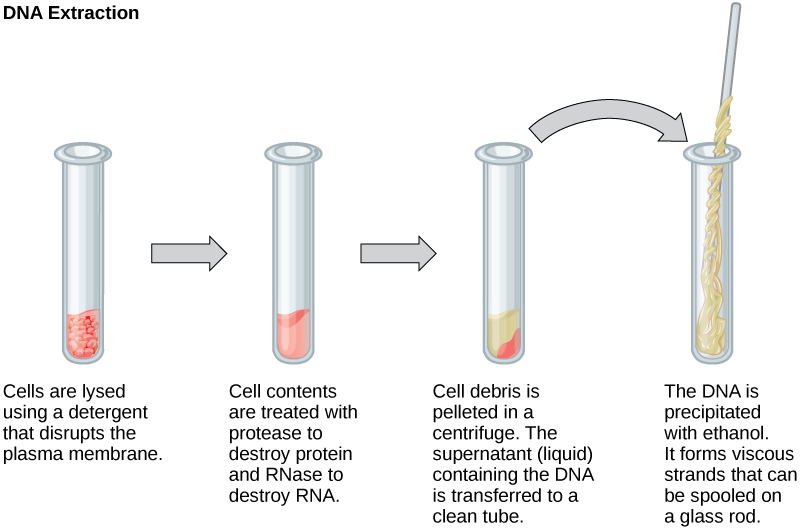
RNA is studied to understand cistron expression patterns in cells. RNA is naturally very unstable because enzymes that break downwards RNA are commonly nowadays in nature. Some are even secreted by our own pare and are very hard to inactivate. Like to Deoxyribonucleic acid extraction, RNA extraction involves the apply of diverse buffers and enzymes to inactivate other macromolecules and preserve only the RNA.
Gel Electrophoresis
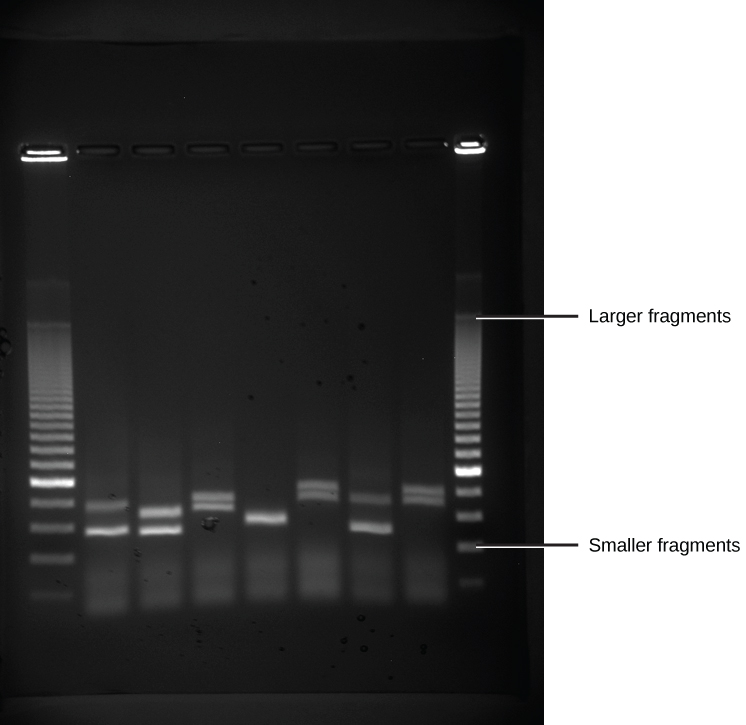
Because nucleic acids are negatively charged ions at neutral or alkaline metal pH in an aqueous environment, they can be moved by an electric field. Gel electrophoresis is a technique used to separate charged molecules on the footing of size and accuse. The nucleic acids tin be separated as whole chromosomes or as fragments. The nucleic acids are loaded into a slot at one cease of a gel matrix, an electrical electric current is applied, and negatively charged molecules are pulled toward the contrary terminate of the gel (the terminate with the positive electrode). Smaller molecules motility through the pores in the gel faster than larger molecules; this difference in the rate of migration separates the fragments on the basis of size. The nucleic acids in a gel matrix are invisible until they are stained with a chemical compound that allows them to be seen, such as a dye. Distinct fragments of nucleic acids appear as bands at specific distances from the top of the gel (the negative electrode terminate) that are based on their size (Figure 10.3). A mixture of many fragments of varying sizes appear equally a long smear, whereas uncut genomic DNA is usually too big to run through the gel and forms a single large band at the pinnacle of the gel.
Polymerase Concatenation Reaction
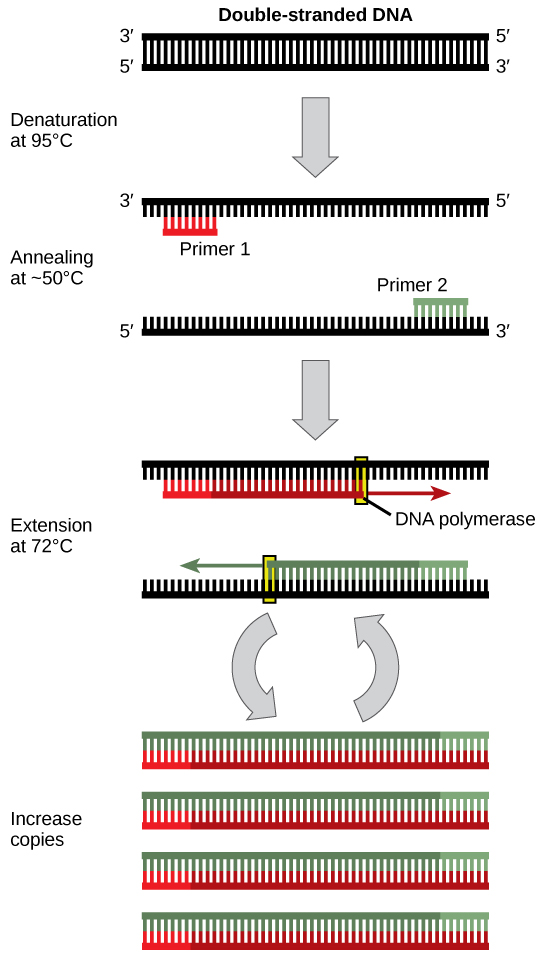
Dna analysis often requires focusing on one or more specific regions of the genome. It also frequently involves situations in which merely 1 or a few copies of a Dna molecule are available for farther assay. These amounts are insufficient for almost procedures, such every bit gel electrophoresis. Polymerase chain reaction (PCR) is a technique used to apace increase the number of copies of specific regions of Dna for further analyses (Figure 10.4). PCR uses a special form of Dna polymerase, the enzyme that replicates Deoxyribonucleic acid, and other short nucleotide sequences called primers that base pair to a specific portion of the Dna existence replicated. PCR is used for many purposes in laboratories. These include: 1) the identification of the owner of a Dna sample left at a crime scene; ii) paternity analysis; iii) the comparison of modest amounts of ancient DNA with modern organisms; and 4) determining the sequence of nucleotides in a specific region.
Cloning
In full general, cloning ways the cosmos of a perfect replica. Typically, the give-and-take is used to describe the creation of a genetically identical re-create. In biology, the re-creation of a whole organism is referred to as "reproductive cloning." Long earlier attempts were made to clone an entire organism, researchers learned how to re-create short stretches of Deoxyribonucleic acid—a procedure that is referred to as molecular cloning.
Molecular Cloning
Cloning allows for the creation of multiple copies of genes, expression of genes, and study of specific genes. To go the Dna fragment into a bacterial jail cell in a form that volition be copied or expressed, the fragment is first inserted into a plasmid. A plasmid (as well called a vector in this context) is a small round Dna molecule that replicates independently of the chromosomal Dna in bacteria. In cloning, the plasmid molecules can exist used to provide a "vehicle" in which to insert a desired DNA fragment. Modified plasmids are usually reintroduced into a bacterial host for replication. As the bacteria divide, they copy their own DNA (including the plasmids). The inserted DNA fragment is copied along with the rest of the bacterial DNA. In a bacterial prison cell, the fragment of DNA from the homo genome (or another organism that is being studied) is referred to as foreign DNA to differentiate information technology from the DNA of the bacterium (the host Dna).
Plasmids occur naturally in bacterial populations (such as Escherichia coli) and have genes that can contribute favorable traits to the organism, such as antibiotic resistance (the ability to be unaffected by antibiotics). Plasmids have been highly engineered as vectors for molecular cloning and for the subsequent large-scale production of important molecules, such as insulin. A valuable characteristic of plasmid vectors is the ease with which a strange Deoxyribonucleic acid fragment tin be introduced. These plasmid vectors comprise many short Deoxyribonucleic acid sequences that can exist cut with dissimilar commonly available restriction enzymes. Brake enzymes (as well called brake endonucleases) recognize specific Deoxyribonucleic acid sequences and cut them in a predictable manner; they are naturally produced by bacteria as a defense mechanism against strange DNA. Many restriction enzymes brand staggered cuts in the two strands of DNA, such that the cut ends take a 2- to 4-nucleotide single-stranded overhang. The sequence that is recognized by the restriction enzyme is a 4- to eight-nucleotide sequence that is a palindrome. Like with a give-and-take palindrome, this means the sequence reads the same forward and backward. In virtually cases, the sequence reads the aforementioned forward on one strand and backward on the complementary strand. When a staggered cutting is made in a sequence like this, the overhangs are complementary (Effigy ten.5).
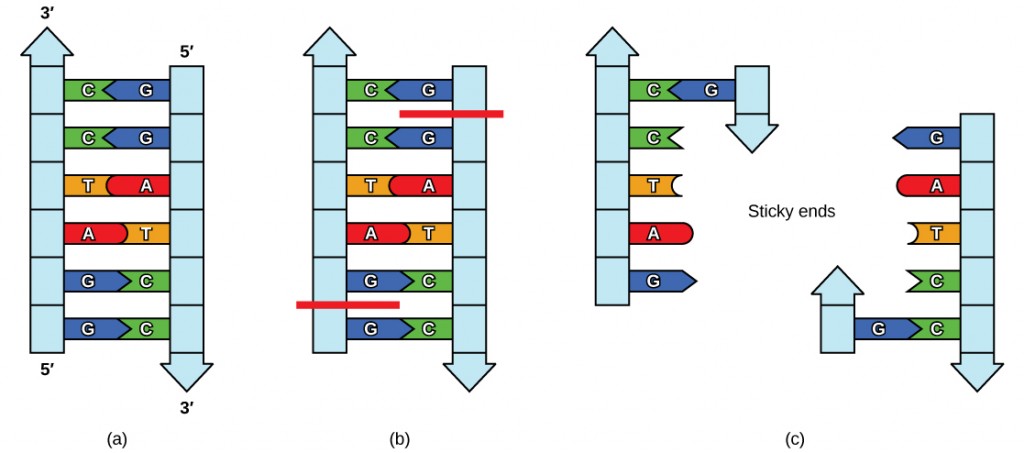
Because these overhangs are capable of coming back together by hydrogen bonding with complementary overhangs on a piece of DNA cut with the same restriction enzyme, these are called "glutinous ends." The procedure of forming hydrogen bonds between complementary sequences on single strands to form double-stranded Deoxyribonucleic acid is called annealing. Improver of an enzyme called Dna ligase, which takes part in Dna replication in cells, permanently joins the Dna fragments when the mucilaginous ends come together. In this style, whatsoever DNA fragment can be spliced between the two ends of a plasmid DNA that has been cut with the same brake enzyme (Figure 10.6).
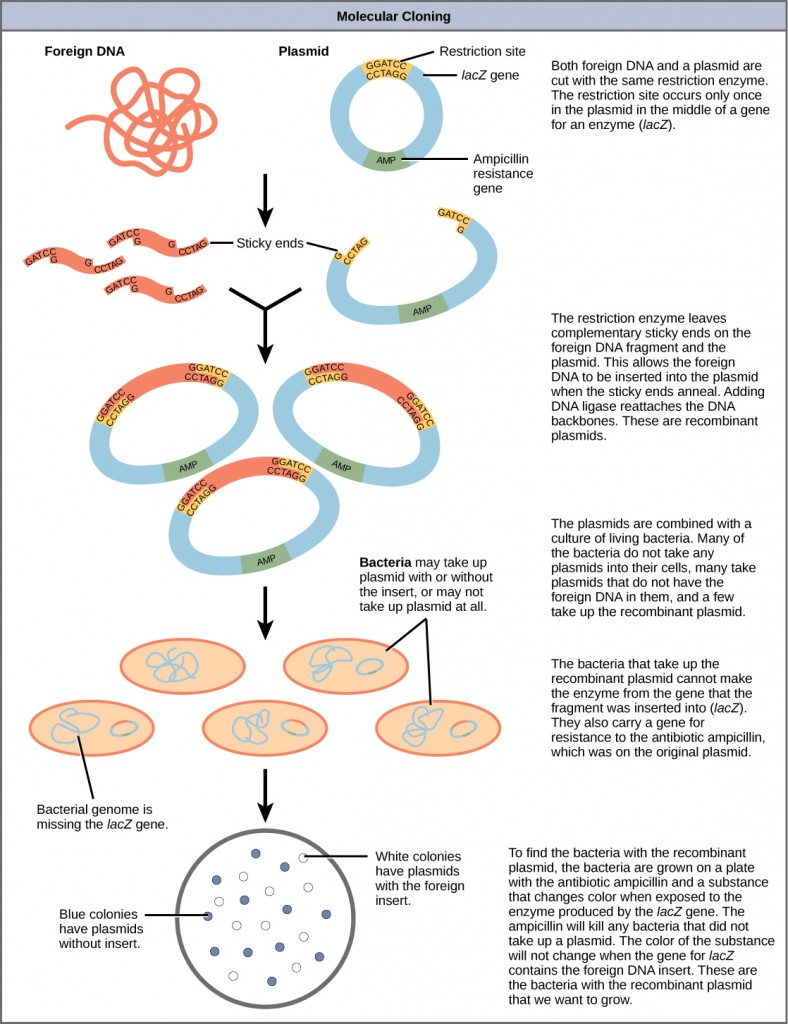
Plasmids with strange DNA inserted into them are called recombinant Dna molecules because they incorporate new combinations of genetic cloth. Proteins that are produced from recombinant DNA molecules are called recombinant proteins. Not all recombinant plasmids are capable of expressing genes. Plasmids may likewise exist engineered to express proteins only when stimulated by certain ecology factors, so that scientists can control the expression of the recombinant proteins.
Reproductive Cloning
Reproductive cloning is a method used to make a clone or an identical re-create of an entire multicellular organism. Almost multicellular organisms undergo reproduction by sexual means, which involves the contribution of DNA from ii individuals (parents), making information technology incommunicable to generate an identical copy or a clone of either parent. Recent advances in biotechnology accept made it possible to reproductively clone mammals in the laboratory.
Natural sexual reproduction involves the wedlock, during fertilization, of a sperm and an egg. Each of these gametes is haploid, meaning they comprise 1 set of chromosomes in their nuclei. The resulting cell, or zygote, is then diploid and contains 2 sets of chromosomes. This cell divides mitotically to produce a multicellular organism. Notwithstanding, the union of just any two cells cannot produce a viable zygote; there are components in the cytoplasm of the egg cell that are essential for the early development of the embryo during its first few jail cell divisions. Without these provisions, there would be no subsequent evolution. Therefore, to produce a new individual, both a diploid genetic complement and an egg cytoplasm are required. The approach to producing an artificially cloned individual is to take the egg cell of one private and to remove the haploid nucleus. And then a diploid nucleus from a body cell of a second individual, the donor, is put into the egg cell. The egg is and then stimulated to split so that development proceeds. This sounds simple, but in fact it takes many attempts before each of the steps is completed successfully.
The commencement cloned agricultural animal was Dolly, a sheep who was born in 1996. The success rate of reproductive cloning at the time was very low. Dolly lived for half dozen years and died of a lung tumor (Figure ten.vii). There was speculation that because the jail cell Dna that gave rising to Dolly came from an older private, the age of the DNA may have affected her life expectancy. Since Dolly, several species of animals (such as horses, bulls, and goats) have been successfully cloned.
There have been attempts at producing cloned man embryos as sources of embryonic stem cells. In the procedure, the DNA from an adult homo is introduced into a human egg cell, which is then stimulated to divide. The technology is similar to the applied science that was used to produce Dolly, but the embryo is never implanted into a surrogate mother. The cells produced are called embryonic stem cells considering they take the capacity to develop into many unlike kinds of cells, such as musculus or nervus cells. The stalk cells could be used to research and ultimately provide therapeutic applications, such as replacing damaged tissues. The benefit of cloning in this example is that the cells used to regenerate new tissues would be a perfect match to the donor of the original DNA. For example, a leukemia patient would non require a sibling with a tissue friction match for a bone-marrow transplant.
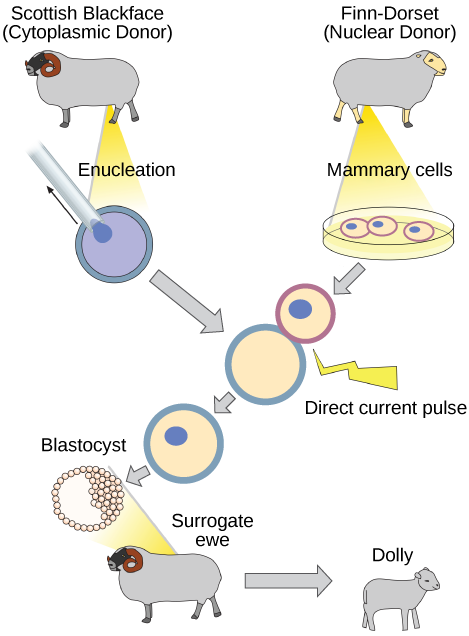
Why was Dolly a Finn-Dorset and not a Scottish Blackface sheep?
Considering even though the original jail cell came from a Scottish Blackface sheep and the surrogate mother was a Scottish Blackface, the DNA came from a Finn-Dorset.
Genetic Engineering
Using recombinant Dna engineering science to alter an organism's DNA to achieve desirable traits is called genetic technology. Addition of strange Dna in the grade of recombinant Deoxyribonucleic acid vectors that are generated by molecular cloning is the most common method of genetic engineering. An organism that receives the recombinant DNA is called a genetically modified organism (GMO). If the strange Dna that is introduced comes from a unlike species, the host organism is called transgenic. Bacteria, plants, and animals accept been genetically modified since the early 1970s for bookish, medical, agricultural, and industrial purposes. These applications will be examined in more detail in the next module.
Concept in Action

Watch this short video explaining how scientists create a transgenic animal.
Although the classic methods of studying the function of genes began with a given phenotype and determined the genetic basis of that phenotype, modern techniques allow researchers to outset at the DNA sequence level and inquire: "What does this gene or DNA element do?" This technique, called opposite genetics, has resulted in reversing the classical genetic methodology. Ane example of this method is coordinating to damaging a body part to determine its function. An insect that loses a wing cannot fly, which means that the wing's function is flight. The classic genetic method compares insects that cannot fly with insects that can wing, and observes that the non-flying insects accept lost wings. Similarly in a opposite genetics arroyo, mutating or deleting genes provides researchers with clues about gene function. Alternately, reverse genetics can exist used to cause a gene to overexpress itself to determine what phenotypic effects may occur.
Department Summary
Nucleic acids can be isolated from cells for the purposes of further analysis by breaking open the cells and enzymatically destroying all other major macromolecules. Fragmented or whole chromosomes tin can be separated on the footing of size by gel electrophoresis. Brusque stretches of Dna can be amplified by PCR. Deoxyribonucleic acid can be cut (and after re-spliced together) using brake enzymes. The molecular and cellular techniques of biotechnology let researchers to genetically engineer organisms, modifying them to accomplish desirable traits.
Cloning may involve cloning minor Deoxyribonucleic acid fragments (molecular cloning), or cloning entire organisms (reproductive cloning). In molecular cloning with bacteria, a desired DNA fragment is inserted into a bacterial plasmid using restriction enzymes and the plasmid is taken upwards by a bacterium, which will so express the foreign Deoxyribonucleic acid. Using other techniques, foreign genes tin exist inserted into eukaryotic organisms. In each case, the organisms are called transgenic organisms. In reproductive cloning, a donor nucleus is put into an enucleated egg cell, which is and so stimulated to carve up and develop into an organism.
In reverse genetics methods, a gene is mutated or removed in some way to identify its event on the phenotype of the whole organism as a mode to determine its function.
Glossary
anneal: in molecular biological science, the process past which two single strands of Dna hydrogen bail at complementary nucleotides to class a double-stranded molecule
biotechnology: the apply of artificial methods to modify the genetic cloth of living organisms or cells to produce novel compounds or to perform new functions
cloning: the production of an verbal copy—specifically, an verbal genetic copy—of a gene, cell, or organism
gel electrophoresis: a technique used to separate molecules on the basis of their ability to migrate through a semisolid gel in response to an electrical electric current
genetic engineering: alteration of the genetic makeup of an organism using the molecular methods of biotechnology
genetically modified organism (GMO): an organism whose genome has been artificially changed
plasmid: a pocket-size round molecule of DNA found in leaner that replicates independently of the main bacterial chromosome; plasmids code for some important traits for bacteria and tin can be used as vectors to transport Dna into bacteria in genetic engineering applications
polymerase chain reaction (PCR): a technique used to make multiple copies of Dna
recombinant Dna: a combination of DNA fragments generated by molecular cloning that does not exist in nature
strong>recombinant poly peptide: a protein that is expressed from recombinant DNA molecules
restriction enzyme: an enzyme that recognizes a specific nucleotide sequence in Dna and cuts the DNA double strand at that recognition site, often with a staggered cut leaving short unmarried strands or "mucilaginous" ends
reverse genetics: a form of genetic analysis that manipulates Deoxyribonucleic acid to disrupt or affect the product of a cistron to analyze the gene's office
reproductive cloning: cloning of unabridged organisms
transgenic: describing an organism that receives Dna from a different species
sullivanhasky1985.blogspot.com
Source: https://opentextbc.ca/biology/chapter/10-1-cloning-and-genetic-engineering/
0 Response to "The Polymerase Chain Reaction (Pcr) Can Be Used to Produce __________ of Copies in a Few Hours."
Post a Comment